
Genetic Management Tools Manual
Michael Raboin, Amanda Vinson, and R. Mark Sharp
April 12, 2020
Source:vignettes/a3manual.Rmd
a3manual.RmdIntroduction
Summary of Major
Functions
Installation
Running Shiny Application
Input
Pedigree Browser
Genetic Value Analysis
Summary Statistics
Breeding Group Formation
ORIP Reporting
Algorithm: Breeding Group
Formation
Algorithm: Genome
Uniqueness
Introduction
The goal of nprcgenekeepr is to implement Genetic Tools for Colony Management. It was initially conceived and developed as a Shiny web application at the Oregon National Primate Research Center (ONPRC) to facilitate some of the analyses they perform regularly. It has been enhanced to have more capability as a Shiny application and to expose the functions so they can be used either interactively or in R scripts.
This work has been supported in part by NIH grants P51 RR13986 to the Southwest National Primate Research Center and P51 OD011092 to the Oregon National Primate Research Center.
At present, the application supports 5 functions:
- Quality control of studbooks contained in text files or Excel workbooks and of pedigrees within LabKey Electronic Health Records (EHR)
- Creation of pedigrees from a lists of animals using the LabKey EHR integration
- Creation and display of an age by sex pyramid plot of the living animals within the designated pedigree
- Generation of Genetic Value Analysis Reports
- Creation of potential breeding groups with and without proscribed sex ratios and defined maximum kinships.
For more information see:
A Practical Approach for Designing Breeding Groups to Maximize Genetic
Diversity in a Large Colony of Captive Rhesus Macaques (Macaca
mulatto) Vinson, A ; Raboin, MJ Journal Of The American
Association For Laboratory Animal Science, 2015 Nov, Vol.54(6),
pp.700-707 [Peer Reviewed Journal]
Summary of Major Functions
Quality Control
Studbooks maintained by breeding colonies generally contain information of varying quality. The quality control functions of the toolkit check to ensure all animals listed as parents have their own line entries, all parents have the appropriate sex listed, no animals are listed as both a sire and a dam, duplicate entries are removed, pedigree generation numbers are added, and all dates are valid dates. In addition, exit dates are added if possible and are consistent with other information such as departure dates and death dates. Current ages of animals that are still alive are added if a database connection is provided via a configuration file and the user has read permission on a LabKey server with the demographic data in an EHR (Electronic Health Record) module. See LabKey documentation.
Parents with ages below a user selected threshold are identified. A minimum parent age in years is set by the user and is used to ensure each parent is at least that age on the birth date of an offspring. The minimum parent age defaults to 2 years. This check is not performed for animals with missing birth dates.
Creation of Pedigree From a List of Potential Breeders and
LabKey
The user can enter a list of focal animals in a CSV file that will be used to create a pedigree containing all direct relative (ancestors and descendants) via the labkey.selectRows function within the Rlabkey package if a database connection is provided via a configuration file and the user has read permission on a LabKey server with the demographic data in an EHR (Electronic Health Record) module.
Two configuration files are needed to use the database features of nprcgenekeepr with LabKey. The first file is named _netrc on Microsoft Windows operating systems and .netrc otherwise, allows the user to authenticate with LabKey through the LabKey API and is fully described by LabKey documentation
The second file is named _nprcgenekeepr_config on
Microsoft Windows operating systems and
.nprcgenekeepr_config otherwise and is the
nprcgenekeepr configuration
file An image of this example configuration file is included as a
data object and can be loaded and viewed with the following lines of R
code in the R console.
Display of an age by sex pyramid plot
Adapted from https://www.thoughtco.com/age-sex-pyramids-and-population-pyramids-1435272 on 20190603. Written by Matt Rosenberg. Updated May 07, 2019.
The most important demographic characteristic of a population is its age-sex structure. Age-sex pyramids (also known as population pyramids) graphically display this information to improve understanding and make comparison easy. The population pyramid sometimes has a distinctive pyramid-like shape when displaying a growing population.
How to Read the Age-Sex Graph
An age-sex pyramid breaks down a population into male and female genders and age ranges. Usually, you’ll find the left side of the pyramid graphing the male population and the right side of the pyramid displaying the female population.
Along the horizontal axis (x-axis) of a population pyramid, the graph displays the population either as a total population of that age or as a percentage of the population at that age. The center of the pyramid starts at zero population and extends out to the left for males and right for females in increasing size, or proportion of the population.
Along the vertical axis (y-axis), age-sex pyramids display two-year age increments, from birth at the bottom to old age at the top.
Genetic Value Analysis Reports
The Genetic Value Analysis is a ranking scheme developed at ONPRC to indicate the relative breeding value of animals in the colony. The scheme uses the mean kinship for each animal to indicate how inter-related it is with the rest of the current breeding colony members. Genome uniqueness is used to provide an indication of whether or not an animal is likely to possess alleles at risk of being lost from the colony. Under the scheme, animals with low mean kinship or high genome uniqueness are ranked more highly.
Breeding Group Formation
One of the goals in breeding group formation is to avoid the potential for mating of closely related animals. Since behavioral concerns and housing constraints will also be taken into account in the group formation process, it is our goal to provide the largest number of animals possible from a list of candidates that can be housed together without risk of consanguineous mating. To that end, this function uses information from the Genetic Value Analysis to search for the largest combinations of animals that can be produced from a list of candidates.
The default options do not consider the sex of individuals when forming the groups, though this has likely been a consideration by the user in selecting the candidate group members. Optionally the user may select to form harem groups, which considers the sex of individuals when forming groups and restricts the number of males to one per group.
Installation
You can install the CRAN version of nprcgenekeepr from the R console prompt with:
install.packages("devtools")
devtools::install_github(file.path("rmsharp", "nprcgenekeepr"))You can install the development version of nprcgenekeepr from GitHub from the R console prompt with:
install.packages("devtools")
devtools::install_github(file.path("rmsharp", "nprcgenekeepr"))All missing dependencies should be automatically installed.
Running Shiny Application
The toolset available within nprcgenekeepr can be used inside standard R scripts. However, it was originally designed to be used within a Shiny application that can be started with:
library(nprcgenekeepr) # nolint: undesirable_function_linter
runGeneKeepR()Input
The Input tab is the starting point for all analyses. The file should be a delimited, regular text file with a header row specifying the columns. The tab provides information on the allowable columns in input files, and how the columns will be used in quality control of the data. Quality control of studbook data occurs automatically upon file upload.
Presently, the only columns required are those specifying the Ego ID, Sire ID, Dam ID, and Sex. The remaining columns listed are optional, but will be used if they are present in the uploaded file. The table of the tab describes how these optional columns will be used. Additionally, the panel on the left of this tab provides options that can be used during the upload and QC process, such as specifying the field separator used in the uploaded file.
During quality control, a flag is added for the current, living population. This flag is generated based on the information columns provided and is fairly specific to how the breeding population is defined at ONPRC. Two of the options for specifying the population of interest can be toggled through this panel, however. Normally, the breeding colony is restricted to Indian-origin, SPF 4 animals. These two restrictions can be turned off by setting the options on this panel.
Additionally, the population of interest can be specified directly in either the input file, or entered on the Pedigree Browser tab.
Pedigree Browser
The Pedigree Browser tab allows the user to view the input data, specify a population to examine, and output the cleaned studbook or trimmed pedigree.
Upon uploading a studbook file, the data goes through the quality control process described on the Input File Format tab. The cleaned version is displayed on this tab. By default, the entire uploaded studbook will be available for viewing on this tab. The first 10 rows will automatically be shown, but this range can be adjusted using the input boxes at the top. The default setting of showing only the first 10 rows is due to the size of the full ONPRC studbook: loading all 32,000 animals can cause the application to be slow.
The tab also contains functionality for trimming the pedigree. By checking the provided box, the studbook that was uploaded will be trimmed down to just the ancestors of the currently specified population. This will remove any lineages that haven’t contributed to the focal group.
As stated above, this tab will also allow the user to directly specify a population to examine. In the top half of the tab, there is an input box to specify the focal set of animals. The population flag can be reset by adding the desired animal IDs to the box. Once the population flag has been set to the desired group of animals, all further analyses will be relative to this group.
Genetic Value Analysis
The Genetic Value Analysis tab provides all of the options needed for producing a genetic value analysis report. The specifics of generating the genetic value analysis report are described on the Genetic Value Analysis and Breeding Group Description tab.
When the analysis is begun, it will generate a genetic value analysis for the currently-specified population in the pedigree. If no population has been specified, the entire pedigree will go into the analysis. This can be problematic, as the function for calculating the pairwise kinship matrix cannot handle large pedigrees. The kinship calculation is known to handle pedigree files containing up to 6000 individuals. It will not, however, handle the whole ONPRC rhesus studbook (~24,000 animals). The exact maximum pedigree size is not currently known and will need to be tested. Due to these problems, the input studbook will automatically be trimmed to the ancestors of the currently-specified population before the genetic value analysis is begun.
The genome uniqueness threshold input box allows the user to specify what constitutes a ‘unique’ allele in the gene-drop simulation. The algorithm description later in this document provides a more in-depth explanation of how the genome uniqueness calculation uses this information. By default, the gene-drop simulation underlying the genome uniqueness calculation considers an individual as unique if no other members of the current population have inherited the same allele during an iteration of the gene-drop. This can be adjusted using the drop-down box to allow up to four other animals to have inherited the allele and still consider it unique.
After the report has been generated, it can be subset to view a specific group of the animals using the text input box. Both the currently-viewed subset and the full report can be exported to a file from here.
Summary Statistics
This tab provides some descriptions of the population being examined after the genetic value analysis has been run. The tab reports the number of known founders, female founders, male founders, founder equivalents, and founder genome equivalents with the first table, which has a single row. The second table has a row for Mean Kinship and a row for Genome Uniqueness. Each row has the Tukey five number summary, which is the minimum, 1st quartile, mean, median, 3rd quartile, and maximum. Lastly, the tab displays histograms and box plots of the distribution of mean kinship coefficients, the distribution of mean kinship coefficient Z-scores, Distribution of genome uniqueness values.
Breeding Group Formation
The last major function of the R-package is to aid in generating breeding groups that avoid inter-animal relatedness. This tab allows you to build a number of breeding groups from a specified list of candidate animals. It also has an option to build a group by adding animals from a list of candidates to a currently-existing group.
In the top half of the tab, there are entry boxes and menus to adjust the options of the analysis. By default, the analysis will ignore relatedness between animals that is more distant than the second cousin level, pairwise relatedness involving an animal under 1 year of age, and relatedness between all females. All of these options can be adjusted before the analysis is run, however.
If the desire is to add animals to an existing group, the IDs of the candidate animals can be entered into the first text box (just as they would be if a new group were being generated). The IDs of the current group members can be added into the second text box. It should be noted that it will cause an error if the provided candidate animals or current group member IDs are not part of the population for which a genetic value analysis was run. The kinship matrix produced for this analysis provides the pairwise kinship values used by the group formation functions.
After the simulation is done, the first group will be displayed automatically. The group being displayed can be changed with the drop-down menu. Whichever group is currently being displayed can then be downloaded with the export button.
Genetic Value Analysis and Breeding Group Formation Description
This tab contains more in-depth descriptions of how the Genetic Value Analysis is created, and how breeding groups are formed by the program.
ORIP Reporting
The ORIP Reporting tab will eventually contain information for reporting to the Office of Research Infrastructure Programs (ORIP). This tab may end up being merged with the Summary Statistics tab and contain a number of statistics, tables and histograms. Alternatively, this may contain a subset of information from the Summary Statistics tab presented as a formatted report that can be exported and submitted to ORIP. The exact information that needs to be submitted for ORIP recordkeeping is still under discussion.
Algorithm: Breeding Group Formation
The group formation process is accomplished by using an algorithm for determining the maximal independent set (MIS). In graph theory, a maximal independent set is the largest set of vertices in a graph where no two share an edge. In breeding group formation, the vertices are animals, and the edges are the kinships that need to be considered. For a given group of animals and pairwise kinships, there are potentially many maximal independent sets, depending on which animals are included or excluded from the final group. In order to effectively sample the set of MISs, we use random selection of animals and repeat the MIS generation numerous times. This allows us to sample a number of MISs and then choose the one that best fits our selection criteria. For our purposes, we want the largest group that can be formed from this set of animals, where none have concerning relatedness to each other.
The algorithm requires several pieces of information:
- The candidate animals
- A matrix of pairwise kinships between candidate animals
- The number of groups desired from the list of candidate
animals
- The number of simulations to run.
* This is equivalent to the number of random MISs to generate and compare.
- Information on which inter-animal relationships (if any) should be ignored.
Data Pre-processing
Before the group formation algorithm begins generating MISs, the data is pre-processed to remove any animals and pairwise kinships that should not be considered.
Specifically:
- The candidate animals provided are checked, and any that were
designated as low-value by the genetic value analysis will be removed
from further consideration.
* This behavior can be toggled off to allow low-value animals in the formation process
- The pairwise kinship data is filtered down to only the kinship
between candidate animals.
- If an age threshold has been set, kinships involving animals below
the threshold will be filtered out.
* This allows the algorithm to ignore young animals, as young animals typically go to whatever social group their dam does.
* By default, we ignore animals under 1 year of age
- Pairwise kinships below the specified level will be filtered
out.
* By default, we ignore relatedness more distant than 2nd cousin
- Pairwise kinships between females will be filtered out
* This allows females of the same matriline to be part of the same group like they would be in the wild.
* This behavior can be toggled off to prevent relatedness between females.
Random Maximum Independent Set Generation
After any animals and relationships that should be ignored are removed from the dataset, the algorithm begins using the remaining animals and kinship information to generate potential groups.
The algorithm proceeds by the following steps:
- For I iterations:
- Generate N empty sets, where N is
the desired number of groups to be created.
- While there are candidate animals remaining:
i. Pick an animal A randomly from the set of candidate animals
ii. Choose a group G randomly from one of the N groups, and assign A to it
iii. Remove animal A from consideration for all N groups
iv. Remove all animals related to A from consideration from for group G
- Score the groups that were generated
i. For our purposes, we calculate the average group size
- If the score of the new groups is higher than groups that were
previously generated, save the new groups.
- Generate N empty sets, where N is
the desired number of groups to be created.
- Return the currently saved groups
- This should be the best groups encountered in I iterations.
Algorithm: Genome Uniqueness
Genome uniqueness is calculated through the use of a gene-drop simulation to estimate how frequently an animal will possess founder alleles not present in other members of the focal population, or present in a specified number or fewer.
The gene-drop simulation used by the web application is a vectorized version and is shown in the figure below. In an un-vectorized version, if 5000 gene-drop simulations are desired for the estimation process, the population had to be iterated over 5000 times. Since each iteration of the gene-drop is independent, the process can be vectorized so that each element of a vector represents 1 iteration of the gene-drop simulation. In the vectorized version, the population is iterated over once, regardless of the number of simulations desired. This drastically reduces the amount of time necessary for the program to run.
Overview
The basic steps of the gene-drop are:
- Each founder is assigned two unique alleles
- For each subsequent generation:
- Assign genotypes to each member of the generation
- For each animal, find the genotypes of the parents, and select
one allele from each parent randomly.
- For each animal, find the genotypes of the parents, and select
- Assign genotypes to each member of the generation
Once every animal has been assigned a genotype by mendelian inheritance tally the number of unique alleles possessed by each member of the focal population. In the case of this algorithm, we do allow the ‘uniqueness’ threshold to be adjusted so that an allele can be considered unique if it is possessed by N or fewer other members of the focal population.
Vectorized Gene-Drop Details
The vectorized gene-drop simulation follows the same basic process described above. The difference is that instead of dropping one allele at a time, and repeating the simulation N times, the vectorized version drops N independent alleles one time.
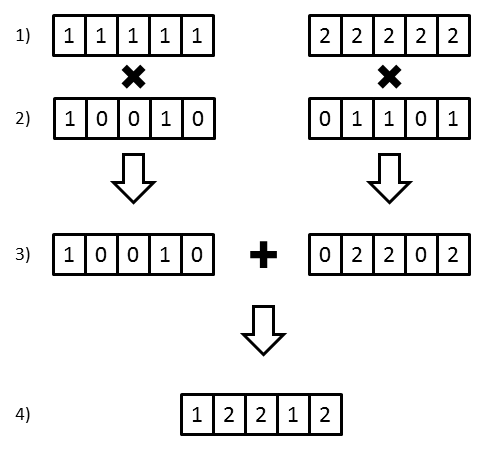
In the vectorized version, each animal has a vector of paternally inherited alleles and a vector of maternally inherited alleles. For each offspring, a random combination of these alleles is produced and dropped down to the offspring by the process below and shown in the following figure:
- To start the simulation, each founder is assigned two unique
founding alleles.
N-element vectors are created of these alleles, where N is the desired number of
simulations. In the example below, this founder was assigned the unique founder
alleles 1 & 2 and 5 simulations were desired.
- Each time alleles need to be dropped from parent to offspring, a
unique
transmission vector is created representing whether or not an allele
was passed to that offspring. The vector is generated to contain a random combination
of 0’s and 1’s. The animal’s paternally inherited alleles are then multiplied by the
transmission vector, while the maternally inherited alleles are multiplied by the
compliment of the transmission vector.
- To generate the final set of alleles received by the offspring, the
maternal and
paternal allele vectors are added together.
- The result is a vector of alleles that this offspring has received from this parent.
Once allele vectors have been generated for every animal in the pedigree, the focal population can be subset out. Within this population of allele vectors, unique alleles can be determined:
For each position on the allele vectors (1:N) - Gather each animal’s two alleles - If the number of other animals possessing that allele is equal to, or below the threshold, score the allele as unique (1) - Otherwise, score the allele as non-unique (0)
Once every position on each animal’s two allele vector’s has been scored, sum all of the scores for an animal and divide by the total number of alleles being considered (2 * number of simulations).
Software Issues
Our goal is to use current R software development practices in an open software environment. Users can see all of the code at github.com/rmsharp/nprcgenekeepr and can submit suggestions and bug reports on our issue tracker at github.com/rmsharp/nprcgenekeepr/issues.
CICD Pipeline Use
The application and associated website is being continuously integrated at each push to the online repository. While often new features being added are not stable or complete, it is uncommon for the application not to run and perform functions that were working before. However, make sure the build was passing by looking for a green R-CMD-check.yaml Passing badge at the top of the README file at https://github.com/rmsharp/nprcgenekeepr/.
Debug Logging
There is a logging system integrated into the package using the package futile.logger. Note the checkbox at the bottom of the side panel on the Input tab. When the Debug on checkbox is checked (it is not checked by default), the application writes to a file named nprcgenekeepr.log in the users home directory. Currently, events occurring the the server.R file are logged as that is where most errors are exposed.
Code Coverage
Code coverage reports are part of the automated build system running in GitHub Actions. We are using the testthat package for unit tests. Currently all code returning values that do not access a database or the file system have coverage with unit tests. Many of these have 100 percent of the lines covered. However, the unit tests are not exhaustive. The practice is to add further tests as errors are detected or when working on the code and a new unit test possibility is discovered. As of 20241223 95.70 percent of the lines are covered.